by Lewis I. Held, Jr.
purchasing information
| Quirks of Human Anatomy by Lewis I. Held, Jr. |  purchasing information |
| back to Quirks index page | |
Figure Legends 4 4.1 * 4.1R * 4.2 * 4.2R * 4.3 * 4.4 * 4.5 * 4.6 * 4.6R * 4.7 figure legends 1* 2 * 3 * 5 * 6 * 7 * A N.B.: An 'R' suffix denotes reflections (commentaries, annotations, and further references) pertaining to the numbered legend that precedes it. [Select any image to enlarge; use back button to return] Fig. 4.1 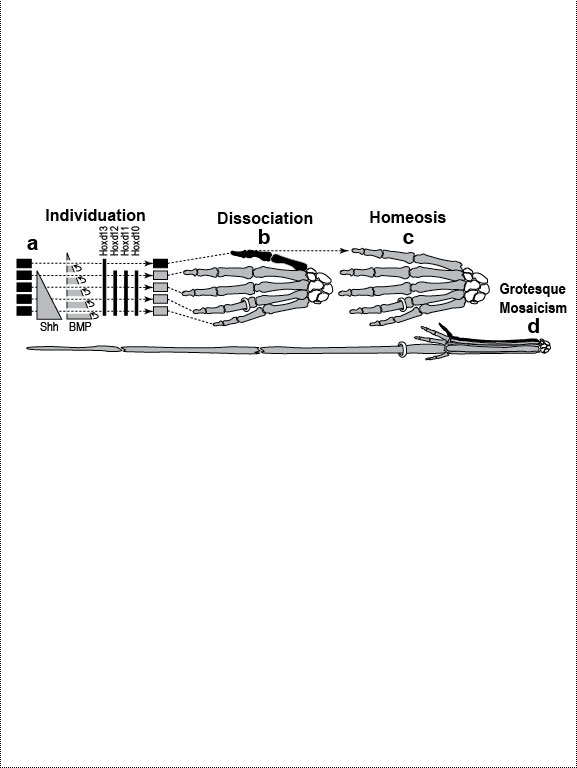 Development (a, b), transformation (c), and fanciful evolution (d) of human fingers. Development (a, b), transformation (c), and fanciful evolution (d) of human fingers.
a. Digits are first detectable as similar condensations of chondrogenic cells (rectangles) [1796] that then become different by "individuation" [833,1638]. This process begins when a signal—Shh (Sonic hedgehog)—diffuses from the pinkie region to form a concentration gradient (triangle) [2193,2252]. Interdigital tissue apparently measures how long it is exposed to Shh [645,1094] and responds by emitting a proportional dose of bone morphogenetic protein (BMP) [1045]. BMP may then activate Hox genes d10-d12 in adjacent digits at a higher threshold than d13. Vertical bars indicate the spatial extent of gene expression along the anterior-posterior axis (not amounts!). Data are from mice [1404]. b. Left hand of a normal adult. The thumb’s unique identity allows it to develop differently ("dissociate") from the other digits. Amazingly, the thumb’s code (d13 ON; d12 OFF) dates back ~400 million years to our fish ancestors [581], who had no bona fide hands or feet [1302]. How the code works is unclear (see text) [379,841,2825,2904], but we do know that it must act indirectly via genes that affect anatomy directly (cf. Fig. 4.2). Embryologically, the thumb is the last digit to form in all land vertebrates except salamanders, where, strangely, it arises first [825]. Our thumb is longer than that of any other primate relative to the index finger [1676], a quirk that enables us to handle tools more adeptly [1848]. c. Left hand of a 35-year-old woman seen at Women’s Clinic University in Graz, Austria, in 1957 [1133], which resembles a bear paw [16]. Her thumb was transformed into a forefinger [2676]—a phenomenon termed "homeosis" (see text)—and the same was true for her right hand, although it also displayed ankylosis of the first metacarpal and trapezium. N.B.: Despite superficial similarities, this case differs significantly from the typical presentation of "triphalangeal thumb" [1033,2582] insofar as (1) all of the muscles and tendons specific to the thumb were missing and (2) opposability was patently lacking [1133]. d. Human left hand redrawn to look like the hand of the pterosaur Anhanguera piscator (which actually lacks our pinkie) [426]. Note the long ring finger, which served as a strut for the pterosaur’s wing membrane [52]. The ability of one member of a meristic series to undergo such drastic divergence ("grotesque mosaicism") should only be possible if it has its own genetic identity (cf. a) [2461]. Fig. 4.1R a. Models of digit patterning are compared in ref. [2547], and models of digit identity are critiqued in refs. [1797,2534]. One unsettled issue concerns thumb identity. The depicted model assumes that thumbness is specified digitally (no pun intended) by a combinatorial code of ON/OFF states ("d13 ON; d10-d12 OFF" [1406,1557]), but an analog mechanism is also possible [641,1794] (i.e., thumbness could be dictated by a lower anterior dose of total Hox protein). Thumb identity may be the universal default state [439,2216] because removal of Shh converts all digits to thumbs [1557]. However, removing Gli3, an Shh transducer that binds Hox proteins [432], evokes odd shapes and more digits [2742]. Another debate concerns the role of BMP paralogs. Lowering the dose of several BMP genes fails to show the threshold effects predicted by the depicted model [131], but the basic idea is still viable because various other Noggin-sensitive (BMP, etc.) genes could be playing a morphogen role [2534]. Finally, there is the nagging question how Shh is transported and interpreted, a broader issue where progress is being made [644,645]. c. The woman’s baby (born later at the same clinic) had the same thumbless 5-finger trait on both hands as she did. Homeosis of their thumbs might be due to broader Hoxd12 expression [1406]. (Hoxd13 mutations in humans can cause syndactyly [1831].) Strangely, however, the feet of mother and baby exhibited preaxial polydactyly (partly duplicated big toes) instead of homeosis. A possibly related syndrome of preaxial polydactyly coupled with triphalangeal thumbs has recently been traced to a defective enhancer of Shh [1033,1525]. d. The white-striped possum also elongated the fourth finger, albeit to a lesser extent [2669] (cf. the fourth toe of the eastern barred bandicoot [758]). The aye-aye converted its middle finger into a long, thin needle for retrieving grubs [52,758], and the slow loris shrank its index finger to a mere stub [1849,1881], while reverting that finger’s nail (atavistically) into a claw for grooming [1260,1371]. Pterosaurs have another remarkable quirk in their hand. In their wrist (not shown) is an elongated carpal ("pteroid") bone that forms a fingerlike strut for the leading edge of the wing [2819,2820]. Another example of such a "wristdigit" is the panda’s "thumb" [578,718] that was so deftly popularized by Stephen Jay Gould [958,961]. Quirks, of course, are in the eye of the beholder. Syndactyly, for example, is considered a dysplasia in humans, but it is the norm for hindfeet in some possum species [758]. Fig. 4.2 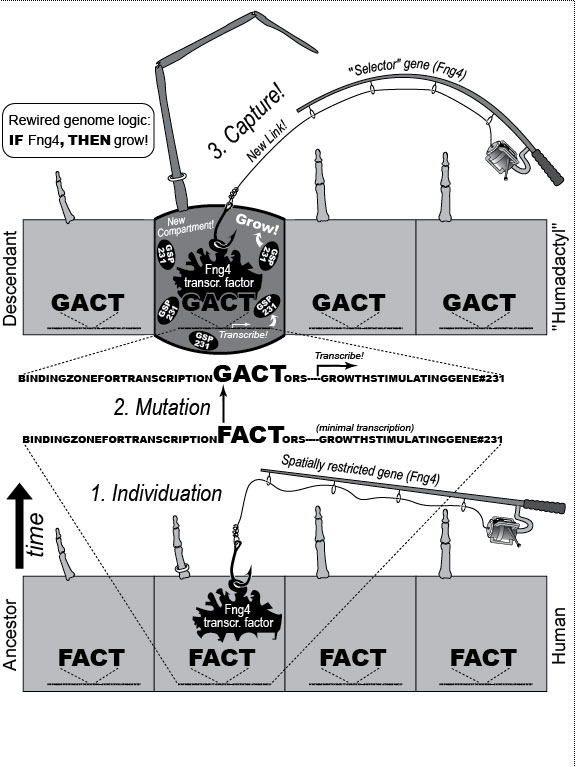 A mental exercise in how a human hand could evolve to resemble a pterodactyl wing. A mental exercise in how a human hand could evolve to resemble a pterodactyl wing.
One major mode of anatomical evolution—possibly the main one [241,401,2204,2478,2838]—involves changes in how genes respond to regional transcription factors [390,2093,2853]. This illustration is based on that mechanism. Each square designates a finger primordium ("phalanx-forming region") [2534], and the bulging square in the upper row denotes a finger growing excessively. Within each square is the DNA sequence (written here in English) of an imaginary gene, #231, that can provoke growth whenever it is transcribed. Normally, however, it is OFF. Step 1. For one finger to grow 10 times longer than the others, it must have a separate identity ("individuation") [833,1582]. In this fanciful scenario, that identity arises when the hypothetical gene Fng4 (fishing pole) has its expression spatially restricted to the ring finger (cf. [1611,2726]), perhaps in response to graded Shh or BMP (cf. Fig. 4.1a). Fng4 encodes the transcription factor Fng4 (bait on the hook). Despite having a unique "area code" at this stage, the finger would look exactly the same because Fng4’s shape doesn’t fit any sequence of letters adjacent to gene #231. Step 2. Over time, sporadic mutations alter the nucleotide sequences in the regulatory zones where transcription factors such as Fng4 can bind [2501,2855]. Those zones are typically a short distance (dashes) from the coding region [1530]. Here, only one letter changes (from F to G). Fig. 4.2R This diagram is a gross oversimplification insofar as (1) proteins can bind DNA not only via a jigsaw-puzzle fit but also by electrical attraction [866], (2) genes can affect other genes at many levels besides transcription [60,61,1178], (3) gene inputs can be negative rather than positive [2620,2817], (4) links can be combinatorial (many to 1) rather than direct (1 to 1) [2414,2637], and (5) fingers are shown as existing before Fng4 expression became restricted, but it could be the other way around [2245,2723]. Indeed, regarding this last point, the tetrapod pattern of Hoxd12/d13 expression is evident in shark fins [816], suggesting that it predated the dawn of digits by ~100 million years (cf. [30]). Step 1 might be rate-limiting, although enhancer-trapping studies show that it can occur simply [401,1639]. Step 2 could also be slow [118,391] because actual binding sites are typically ~5 to 10 letters long [22,866,2642], but mutations need not affect just one letter at a time [2814,2851]. Cut-and-paste shuffling of "word" and "sentence" fragments [329,2130] through transposition, gene conversion, and unequal crossing over [676] refutes the old "monkeys-on-typewriters" canard that evolution can’t create complexity [588,960,1167]. After such rewiring occurs in an individual, the novelty can linger as a polymorphism and, if sufficiently adaptive, spread and eventually saturate the population [185,2851,2855]. Case studies that illuminate various aspects of the depicted scenario include (1) the accrual of binding sites throughout the genome after a selector gene arises (mammalian limb [2701]); (2) the evolutionary flux of transcription factor binding sites (yeast [246], Drosophila [1814,2632], and in general [1605,2130,2501]); (3) the tolerance of genomic networks for tinkering with their links (bacteria [181,1249]); and (4) the instrumental involvement of rewiring in evolution (reviews [1255,1605]). The most relevant case may be the amazing tale of how the swordtail fish got its sword, a question Darwin himself contemplated in the second half of The Descent of Man, and Selection in Relation to Sex [708]. Recent revelations from evo-devo research have essentially solved the mystery of how one element of a meristic series can be singled out for extraordinary growth [390,2263]. If, at some future date, a mutation (not shown) were to disable Fng4, then it would, in this analogy, break the pole and release all the "fish" that had been hooked over the eons [2815]. In that case, the ring finger would homeotically revert to its mundane morphology. Reversions to prior evolutionary states are termed "atavisms" [1056,2067,2482]. Atavisms can involve reappearance of lost structures in rare individuals or in whole species [151,1053,2178,2482]. Atavisms in humans putatively include apelike fur [153,765,862], doglike nipples along the milk lines [408], and fishlike "gill slits" on the neck [666]. Instances in nonhuman species include bird teeth [1100], snake fingers [268], horse toes [963], frog tails [1077], whale hindlegs [1053], and fly hindwings [818,860]. Traits that have reestablished themselves in whole species include lizard toes [1415], larval stages in salamanders [444], shell-coiling in gastropods [492], wings in stick insects [2497,2799], spots on fly wings [2094,2852], and sex combs on fly legs [1425]. Taxon-level reversals could theoretically start with "hopeful monster" mutants [661,662,957,2179,2733] or arise through a gradual awakening of dormant pathways [1040,2475]. However, dormant circuits are difficult to resuscitate after more than ~10 million years [310,311,1825,2584,2585] because of deterioration from disuse [1659,2922]. Fig. 4.3 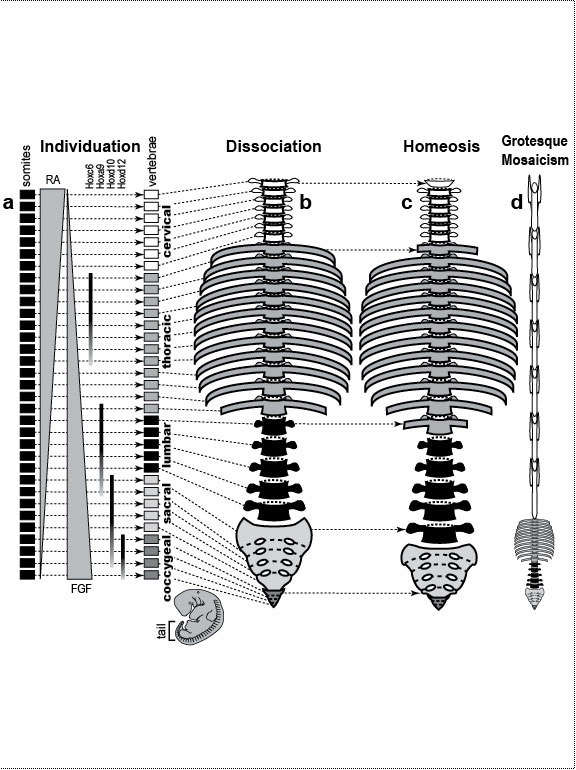 Development (a, b), transformation (c), and fanciful evolution (d) of human vertebrae. Development (a, b), transformation (c), and fanciful evolution (d) of human vertebrae.a. Vertebrae (and ribs [1218,1928]) develop from blocks of tissue called "somites" (rectangles). Somites and vertebrae are slightly out of phase [82,1810] (not shown; cf. [23] for an even stranger shift in frogs). Occipital somites are omitted because they form part of the skull [1211,1307,1592]. Somites arise in a wave from head to tail and initially look alike [123]. They acquire separate identities ("individuation") through a Hox code [521,791,2785] that uses a subset of our 39 Hox genes [1011]. Gradients (triangles) of retinoic acid (RA) [1621] and fibroblast growth factor (FGF) [684] specify where Hox genes get turned ON [1483], and these ON/OFF states endure [1379]. Graded vertical bars denote expression zones for genes whose anterior limits coincide with type transitions [318,376,1230], although expression in presomitic mesoderm is more critical [371]. Limb buds (not shown) may also be positioned by RA/FGF because tadpole tails grow legs (and pelvic girdles) when exposed to RA [2234]! The ground state for vertebrae is thought to be "thoracic" (i.e., rib bearing) [2785]. Below is a 31-day-old human embryo [2311]. Note the somites (along the spine) and the tail (which stops growing later to form the coccyx and sacrum) [1076]. Rarely, humans are born with short, external tails [91,561,2178,2802], but these cases may not be true atavisms [2683]. The specific vertebrae that comprise the sacrum vary among individuals [2806]. b. Axial skeleton of a normal adult (ventral perspective; i.e., ribs curving toward viewer). Different types of vertebrae are indicated by different shadings. c. Axial skeleton (ventral perspective) from a man’s body donated ca 1882 to Amsterdam’s Vrolik Museum [1938]. The most anterior vertebra of every region appears to be shifted in identity by one unit. (The depicted shift of first cervical to an occipital identity (i.e., base of the skull) is conjectural because his skull was not retained.) Similar shifts are seen in Gbx2, Gdf11, or Cdx mutant mice [371,834] or in embryos treated with RA [499,1368]. ("Frame shifting" of identity relative to merism also occurs in digits [2718].) Asymmetries in rib formation (here first thoracic and first lumbar) are surprisingly common in human axial homeoses [845] and may be due to insufficient RA [276]. More drastic homeoses (e.g., ribs eliminated [371] or ribs everywhere [2785]) occur when Hox genes are broadly misexpressed or collectively disabled [1707]. d. Axial skeleton of a human redrawn to resemble that of the pterosaur Quetzalcoatlus northropi [426]. Note the incredibly elongated cervical vertebrae (dwarfing a giraffe’s neck [2425]). The opposite trend (cervical compression) is seen in whales [188]. We do not know why the number of cervical vertebrae is so constrained in some groups of animals (e.g., pterosaurs [426] and mammals [1850]) but not in others (e.g., sauropods [746] and birds [1873]) [840]. Revamped vertebrae are also seen in turtles, whose ribs fuse with their shell [1842,2179], and in extinct gliding lizards, whose ribs formed the only wings that did not evolve from arms [2373,2752]. The pterosaur's long neck is counterbalanced by long legs (not shown). Fig. 4.4 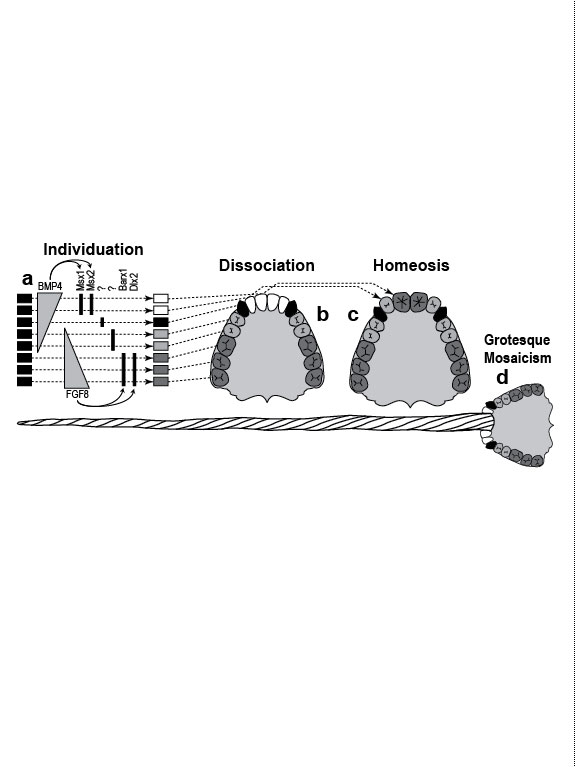 Development (a, b), transformation (c), and fanciful evolution (d) of human teeth. Development (a, b), transformation (c), and fanciful evolution (d) of human teeth.a. Our teeth come from tooth buds (rectangles) [1917,2643]. Identities are dictated by antagonistic gradients of morphogens (BMP4, FGF8) [1561,1770] that turn ON genes for homeobox transcription factors (Msx1 and 2, Barx1, Dlx2, etc.) at different thresholds [2643]. Vertical bars denote expression zones. We do not yet know the codes for premolars and incisors [1346,1695]. Tooth positions may be set by similar circuitry [1867,2059]. The core of each tooth comes from neural crest [412,1743], which imposes its final identity on the overlying, enamel-making epithelium [476,567] through a reciprocal dialogue [413]. Our reptile ancestors had only peglike teeth [1944], so the human default dentition may likewise be a mouthful of conically shaped teeth resembling our canines. b. Upper dentition of a normal adult [2027,2374]. (As children, we have no premolars, and our juvenile molars are replaced inexplicably by adult premolars [1866].) All of our teeth flank the midline, and the same is true for other mammals [151], except for one odd bat species (on an island off the African coast) that has three incisors in its lower jaw, the middle one straddling the midline [1321]. Median incisors are rare in other species [151], and they are diagnostic of a mild form of human holoprosencephaly syndrome (cf. Ch. 3) [857]. c. Upper dentition of a 13-year-old girl examined at the University of Minnesota ca 1990 [1332]. Her central incisors looked like molars (with extra cusps), and her lateral incisors like premolars. Her bottom teeth (not shown) were relatively normal (as was her face), but one premolar was missing. She had no family history of dental defects, so the genetic basis is unclear, although she did show congenital hearing loss. d. Upper teeth of a human redrawn to simulate that of the narwhal Monodon monoceros, in which the inner left incisor elongates enormously (in males) to form a tusk [2146]. Overgrowth might occur through targeted overproduction of Shh [566]. Adult narwhals actually lack all teeth except the tusk. Fig. 4.5 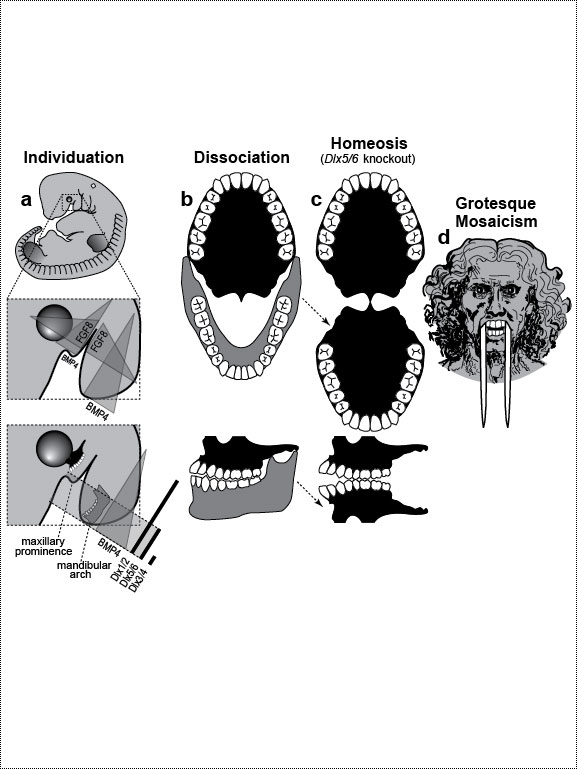 Development (a, b), transformation (c), and fanciful evolution (d) of the human jaw (upper vs. lower). Development (a, b), transformation (c), and fanciful evolution (d) of the human jaw (upper vs. lower).a. Above is a 31-day-old human embryo [2311]. Below are enlarged cartoons of the oral region. The large circle denotes the incipient eye. Our upper jaw comes from the "maxillary prominence" [409,1509], whereas our lower jaw comes from the "mandibular arch" [1142]. The oral area is criss-crossed by gradients of the same morphogens that cause teeth individuation (cf. Fig. 4.4). FGF8 spreads outward from the gap between the maxillary and mandibular areas [477,1123], whereas BMP4 diffuses inward from the tips of the two bumps [2172,2364]. Together they coax Dlx genes into a nested pattern, although Dlx enhancers studied thus far respond counterintuitively (positively to FGF8 and negatively to BMP4) [1992]. The nested domains (slanted bars) act as a "Dlx code" that distinguishes upper (Dlx1/2 ON; Dlx5/6 OFF) versus lower (Dlx1/2 ON; Dlx5/6 ON) jaw [636], where the shorthand "Dlx1/2" means "Dlx1 and Dlx2." The maxillary is defined by Dlx5/6 OFF, whereas the mandibular is defined by Dlx5/6 ON [1292]. This Dlx code may have existed for eons in our fish forebears [2487] before fortuitous mutations caused it to capture anatomy-affecting genes (cf. Fig. 4.2), which led to visible dissociation of upper versus lower dentitions (b) [299]. b. Dentition of an adult human (upper jaw above, lower below). Note the differences (upper vs. lower) in sizes of incisors and the cusps of molars [1004]. Note also the occlusion of alternating cusps from the two surfaces (side view below) [1573,2154], which enables us to chew more effectively [50,2768]. Such a precise fit may only have been possible after our mammal ancestors stopped replacing teeth continuously (like reptiles) and started making two sets: deciduous (baby) teeth and permanent (adult) teeth [59]. c. Transformation of lower into upper jaw due to inactivation of Dlx5/6 in mice [189,633], rendered here in human form. The same phenotype is obtained by blocking Endothelin-1 signaling [1961,2239], which acts upstream of Dlx5/6 [2171]. (Disabling of Activin signaling also targets the lower jaw but does not transform it [1695].) d. Teeth of a human redrawn to mimic the saber-toothed cat Smilodon lethalis [458,1098], where the upper jaw has huge canines, but the lower jaw does not. Fig. 4.5R a. Man’s likeness to a fish at this stage may have been why Darwin decided to use such a drawing as his first figure in Descent of Man because no other image so clearly conveys his book’s theme as stated at its end: "Man still bears in his bodily frame the indelible stamp of his lowly origin." Gill slits persist atavistically as cervical fistulae in rare humans [91,666,2178], although such "humafish" would presumably still need scuba gear to respire effectively underwater. b. How mammals make a certain number of teeth [11] and how each tooth makes a definite number of cusps [1299] are active areas of investigation in evo-devo [1346,1770]. d. Other examples of upper-lower dental divergence include the tusks of elephants, walruses, boars, and narwhals (cf. Fig. 2.4) [52,2178] and the fangs of snakes [2897]. The evo-devo story of how the snake got its fangs has recently been deciphered [2705]. One knotty question that evo-devo is starting to address is why certain teeth grow continuously (e.g., rodent incisors) but most do not [1769,2590]. Fig. 4.6 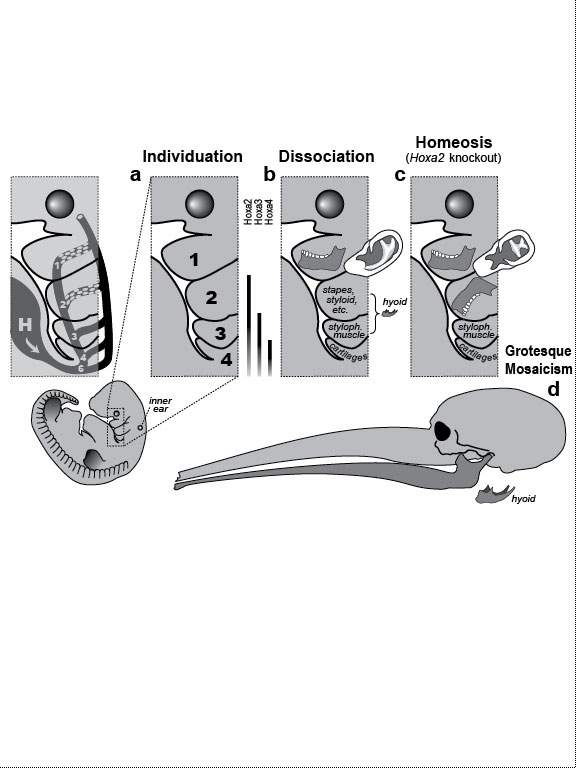 Development (a, b), transformation (c), and fanciful evolution (d) of the branchial arches. Development (a, b), transformation (c), and fanciful evolution (d) of the branchial arches.a.. Human embryo at 31 days postfertilization with neck region magnified to show branchial arches (BA1-4), branchial clefts (grooves between the arches), and underlying aortic arches (cartoon at left) that convey blood (arrow) from the heart (H) [2311]. Numbering obeys phyletic convention. BA5 never forms in humans [143], nor do the clefts perforate as in fish, where they would form gill slits. Aortic arches 1 and 2 disintegrate (schematized as Swiss-cheese holes) shortly after they arise and individuate (cf. [43] for variations). This strategy of making structures and then destroying them is idiotic from an engineering standpoint but makes sense evolutionarily, given that plumbing, in general, is easier to revamp than to reinvent [1838]. Darkly shaded protrusions are limb buds. The large circle is the eye primordium. b. Some BA derivatives [2295,2311]. BA1 makes our lower jaw, BA2 our styloid process, BA3 our stylopharyngeal muscle, and BA4 our laryngeal cartilages. Our outer ear arises from BA1 and BA2, as do our middle ear bones (stapes from BA2; others from BA1) [91,757]. Our inner ear starts dorsally (a) as an otic pit that sinks to form a separately developing vesicle [229] in accordance with its own (ancient) ancestry [2513], although, interestingly, it also requires Dlx5/6 [2201]. Our hyoid bone comes from BA2&3. c. Transformation of BA2 to BA1 due to inactivation of Hoxa2 in mice [880,2180], rendered here in human form. A second lower jaw forms in mirror image to the normal one [2420]. Mirror duplication of the front part of the outer ear is inferred, given that approximately the front one third comes from BA1 and the hind two thirds from BA2 [719]. Absence of hyoid (part of which comes from BA2) was documented in frogs [130]. d. Head of a human redrawn to approximate the oversized snout of the giant anteater Myrmecophaga tridactyla [1669]—a feature also found in cetaceans (cf. [908]; their Fig. 23.18). The opposite disjunction (hyoid vs. jaw size) occurs in male howler monkeys [1161,1162], monitor lizards [2527], and predatory frog tadpoles [615]. Fig. 4.6R a. Development of the aorta (not shown) involves unilateral destruction of an aortic arch [2876]. Interestingly, we make an aorta from our left fourth arch and erase our right one [2311], whereas birds do the opposite [1838]. Because mammals and birds both evolved from reptiles, in which arches stay nearly symmetrical, we are left to ponder why the left was commandeered in one clade and the right in another [1488,1838]. It is not known whether the nested domains of Hox gene expression (vertical bars [1936]) are delimited by a morphogen gradient(s) [990]. The domains may provide a code like that for vertebrae (cf. Fig. 4.3) [995]. Fig. 4.7 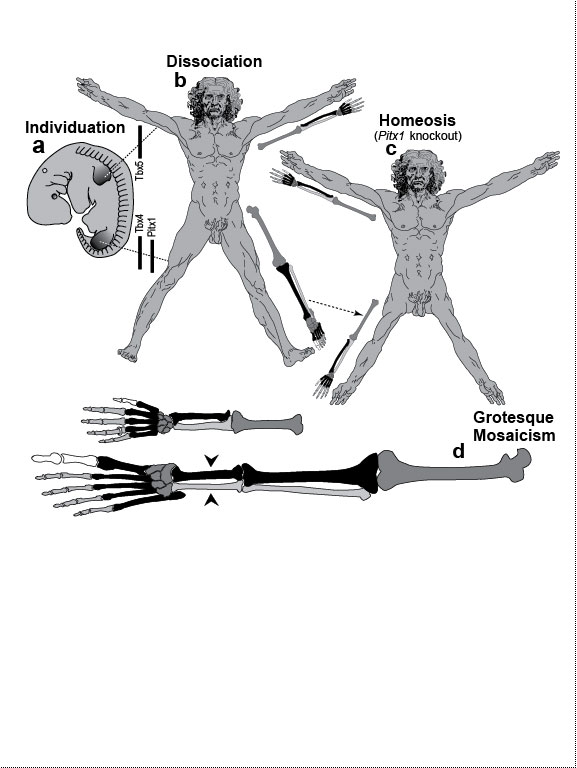 Development (a, b), transformation (c), and fanciful evolution (d) of the arm and leg. Development (a, b), transformation (c), and fanciful evolution (d) of the arm and leg.a. Human embryo at 31 days postfertilization, showing limb buds (shaded protrusions) [1888,2311]. Vertical bars designate genes expressed in each respective limb bud [2568]. In chick embryos, forelimb versus hindlimb identities can be switched by misexpressing Pitx1 [1567], Tbx4, or Tbx5 [2205,2559]. In mice, Pitx1 is also effective [623], but Tbx4 and Tbx5 are not [1113,1765,1844]—a phyletic difference that is not yet understood [1208,1728,2503]. Hox genes along the body axis are also involved in assigning identities (cf. Fig. 4.3) [318,1902,2258], although the exact code is unclear [623]. Many other genes are expressed differently in arm versus leg [1245,2382], some of which are undoubtedly downstream effectors. b. Normal human anatomy. The skeletal similarity of the arm and leg—indeed, of all vertebrate limbs—bolstered Darwin’s case for common descent [559]: "What can be more curious than that the hand of a man, formed for grasping, that of a mole for digging, the leg of the horse, the paddle of the porpoise, and the wing of the bat, should all be constructed on the same pattern, and should include the same bones, in the same relative positions?" In other primates, the feet are more handlike in both structure and function [2321]. c. Transformation of leg into arm due to inactivation of Pitx1 in mice [2540], rendered here in human form. In fact, the observed homeosis is only partial, and no 4-armed humans exist [1638]. However, several families of people who walk on all fours have recently been discovered in Turkey [2564]. d. Human limb bones (arm above; leg below) redrawn to imitate those of the American bullfrog Rana catesbeiana [1078]. Arrowheads indicate the extra tibia ("tibiale") and fibula ("fibulare") that characterize all extant frog species [381,2633]. This ability of frogs—and a few primates [715,873]—to convert ankle bones into long bones refutes the cliché [2503] that tetrapod limbs have an immutable (4-zone) bone formula along their length—viz., "1:2:many:≤5," where "≤5" denotes the number of digits. This heretical "magic trick" may not have been as hard as it seems: notwithstanding their later disparity, the rudiments of ankle bones are the same size as the long bones when they first arise [1622,2526], but they then stop growing in most vertebrates [2844]. Hence, frogs might have lengthened them by merely unleashing their inherent growth potential. top of page
Lewis I. Held, Jr. is Associate Professor in the Department of Biology at Texas Tech University.
|
 © 2009 Thomas B. Brody, Ph.D. |