Select image to enlarge
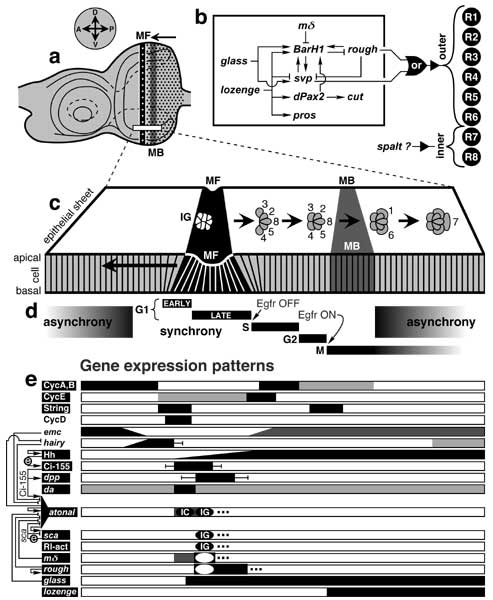
Figure 7.9
Genetic logic of eye development and dorsal-ventral stripes of gene expression in the eye disc epithelium during the 3rd instar and early pupal period.
See also App. 7.
a. Mature left eye disc. Vertical stripes are the morphogenetic furrow (MF) and mitotic band (MB). Compass (above) gives axes in the fate map (cf. Fig. 7.1). The MF traverses the epithelium from P to A, leaving a hexagonal lattice of nascent ommatidia (dots) in its wake. The MB is needed to supply enough cells to finish assembling the ommatidia [1003, 3655, 4712].
b. Circuitry among genes that encode transcription factors for photoreceptor cell fates (—> activation; —| inhibition; see text). Abbreviations: pros (prospero), svp (seven-up). BarH1 is one of two genes (the other being BarH2) in the Bar Complex. A combinatorial code for R cells must exist [1272], though there is little evidence for it except for rough and svp, which dictate outer-type identity, and even there the data is murky. Neither rough nor svp can convert R7p to an outer state in a sevnull background [232, 1856], so R7p must require the Boss-Sev reaction to become neural [4551]. Also, Rough and Svp are not interchangeable: they have different effects when expressed in cone cells [264, 4551].
c. Imaginary slice of the columnar epithelium (magnified from box in a) at one instant of time. The MF is a groove (black) that runs along the D-V axis. It moves from P to A (long arrow). Each nascent ommatidium that it creates undergoes a series of transformations (short arrows). These stages can be seen in the (static) A-to-P sequence of nodes along each row because each successive column is older. The 'intermediate group' stage (IG) precedes the rosette stage (not shown; cf. Fig. 7.5) [2042]. Within each IG, 2-3 R8p-like cells express senseless (not shown) [1279], which then turns OFF in all but one cell that becomes R8. Ovals (top view) are apical profiles of single cells (mystery cells not shown). Vertical lines (side view) are cell boundaries. The epithelium is actually thicker, and cell packing is less regular [4715].
d. Stages of the cell cycle. The span of each stage along the A-P axis is indicated by a black bar. As cells enter the furrow, they synchronize but lose this synchrony around the time that they begin mitosis [4715]. Arrows mean that a cell's EGFR pathway must be OFF in order for it to go from G1 to S but must be ON for it to go from G2 to M [185]. String is the downstream gating factor for the latter transition [185].
e. Domains of gene or protein expression relative to eye epithelium in c. Each stripe of gene expression along the D-V axis (parallel to the MF) is shown here in terms of its span along the A-P axis. Degrees of gene expression are indicated by shades of gray or by graded slopes. If LOF effects have been found, then the name of the gene/protein is in white letters on a black rectangle. Error bars indicate confidence limits. Black ovals denote expression in IGs or earlier 'initial clusters' (ICs) [4208], while a black frame (hollow oval) denotes expression between IGs. Thus, sca exhibits a punctate pattern of spaced islands (IGs), while atonal displays a wider stripe with intense A (IC) and P (IG) foci with a P edge that describes a sine wave [183, 184, 1921, 2461]. ICs and IGs are out of phase relative to one another (not shown), and the IC phase is dispensable [1616]. Genes or proteins: atonal [1077, 2493]Δ (autoregulation [4208]Δ; links to glass [2042], hairy [2042]Δ and sca [1077]), Ci-155 (the activator form of Cubitus Interruptus) [1077, 1616]Δ, CycA-E (Cyclins A-E) [185]Δ, da (daughterless) [506]Δ, dpp (decapentaplegic)[725, 3380], emc (extramacrochaetae) [508], glass [2569]Δ, hairy [2540]Δ, Hh (Hedgehog) [287] (autoregulation [1786, 3238]), lozenge [639]Δ, mΔ [2549]Δ, Rl-act (activated Rolled; a.k.a. MAP kinase [332]) [2493]Δ, rough [725]Δ, sca (scabrous) [183]Δ, sens (senseless) [2540], String [185]Δ.
Wiring at left (—> activation; —| inhibition; circled 'e' = extracellular signal) shows some genetic interactions (see text or references above).
Links in panel b are discussed in the text (cf. Table 7.1). Panel c is adapted from [4715], and d is based on [185, 4291, 4293]. Panel e is styled after [1786, 1788, 4387] with additional data from the above references. For further circuit analysis see [395, 4387], for circuits that affect non-optic head structures see [80, 3380, 3663-3665, 4654], and for comparable analyses in other systems see [309, 2093, 4289].
|
|
