Select image to enlarge
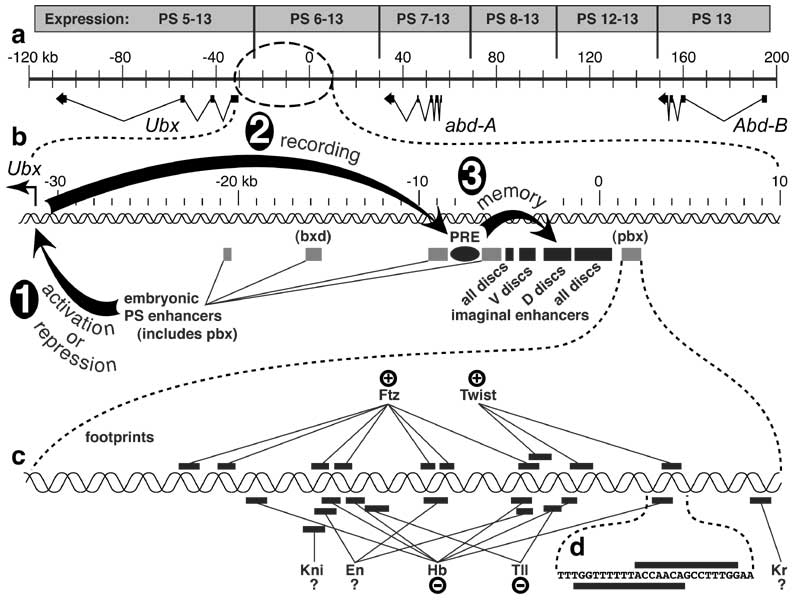
Figure 8.1
The Bithorax Complex (BX-C) and its regulation during development. See also App. 7.
a. The BX-C contains 3 protein-encoding homeotic genes: Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B) [3740]. Exons are drawn as black rectangles, with introns indicated as thin (kinked) connecting lines. When reporter genes (e.g., lacZ) insert into this 320 kb region, their expression is confined to a specific span of parasegments (PS, above) [284, 2773] due to nearby enhancers in the BX-C (cf. Fig. 4.2 for PS numbering) [2987, 3480, 3481, 3940, 4864]. The more rightward the reporter's insertion point (= distal on the right arm of the 3rd chromosome), the more the expression domain recedes toward PS13. This colinearity of enhancers (proximal to distal) relative to body regions (anterior to posterior) is also seen in the order of 'iab' genetic control elements (not shown) [416, 699, 907, 3738]. The reason for the colinearity is unknown [162, 686, 3511, 4152, 4328, 4498], though clever guesses have been offered [1113, 1400, 2191, 2295, 2508, 2672, 3494]. A similar 'Homeobox Homunculus Mystery' [1417, 1805] exists for Hox gene complexes of vertebrates [1111, 1112, 2266, 2292, 4446] -- i.e., why are Hox genes and enhancers arranged like the parts of a miniature man or fly?
b. Enlarged view of the 40 kb section upstream of Ubx. Three stages in regulation are depicted. In Step 1, embryonic enhancers (shaded rectangles) turn Ubx ON or OFF in specific parasegments based on input that they receive from segmentation genes (c; cf. Fig. 4.2). In Step 2, the PRE (Polycomb Response Element, black oval) indelibly 'records' the cell's state by recruiting activator (Trithorax-Group) or repressor (Polycomb-Group) proteins [3196], depending on whether Ubx is actively transcribed (proteins not shown). Finally, in Step 3, the recruited proteins affect the imaginal disc enhancers (black rectangles) by blocking or aiding nucleosomal silencing of the DNA within > 10 kb. Among the disc enhancers, two drive expression in all discs, one affects mainly ventral (leg) discs, and the other mainly dorsal (wing and haltere) discs.
c. Magnified view of the pbx embryonic enhancer (619 b.p.), showing footprints (bars) of various regulatory proteins. Abbreviations: En (Engrailed), Ftz (Fushi tarazu), Hb (Hunchback), Kni (Knirps), Kr (Krüppel), Tll (Tailless). All of these proteins participate in ectodermal segmentation (cf. Fig. 4.2 and [3680] for tailless, a terminal gap gene) except Twist, a bHLH transcription factor that activates mesodermal target genes [249, 1985, 2842, 4818] in the embryo's D-V patterning system [3576, 4290]. Plus signs indicate activation, and minus signs denote repression, though individual footprint sites have not been tested for function in vivo [3397]. Kr and Kni can repress BX-C genes [680, 3902], but expression of a pbx-driven lacZ reporter is normal in KrLOF or kniLOF embryos [4864] -- arguing against pbx-mediated regulation of Ubx by these agents. En is a repressor for Ubx in the haltere disc [1138, 1162, 1635, 4229], but in the embryo it can behave either as an activator [3481] or repressor [684, 2670, 2725].
d. Nucleotide sequence in an enlarged section the pbx cis-enhancer where a Twist binding site (bar above) appears to overlap a Hb binding site (bar below). It is important to remember that such sites are delimited by DNase protection assays, so the actual contact area with DNA may be smaller. Hence, these proteins might be able to bind simultaneously (i.e., without competing).
Demarcations of PS expression zones in a are only crude estimates due to a limited number of insertion sites [284]. Exons for BX-C genes are after [284, 2773, 3156, 3902], though each of the three genes has other splicing isoforms. Nevertheless, for Ubx at least, the isoforms act alike with regard to specifying parasegment identity [684]. Map coordinates obey convention [283]. Schematic in b is adapted from [3397]. See [3667, 4319] for finer dissections that identify separate Pc-G and Trx-G binding sites. Data in c and d are from [3397, 4864].
|
|

